| Merging nucleus datasets by correlation-based cross-training |
| Wenhua Zhang*1,2, Jun Zhang*2, Sen Yang2, Xiyue Wang2, Wei Yang2, Junzhou Huang2, Wenping Wang#1,3, Xiao Han#2 |
| 1The University of Hong Kong, 2Tencent AI Lab, 3Texas A&M University, |
| |
 |
|
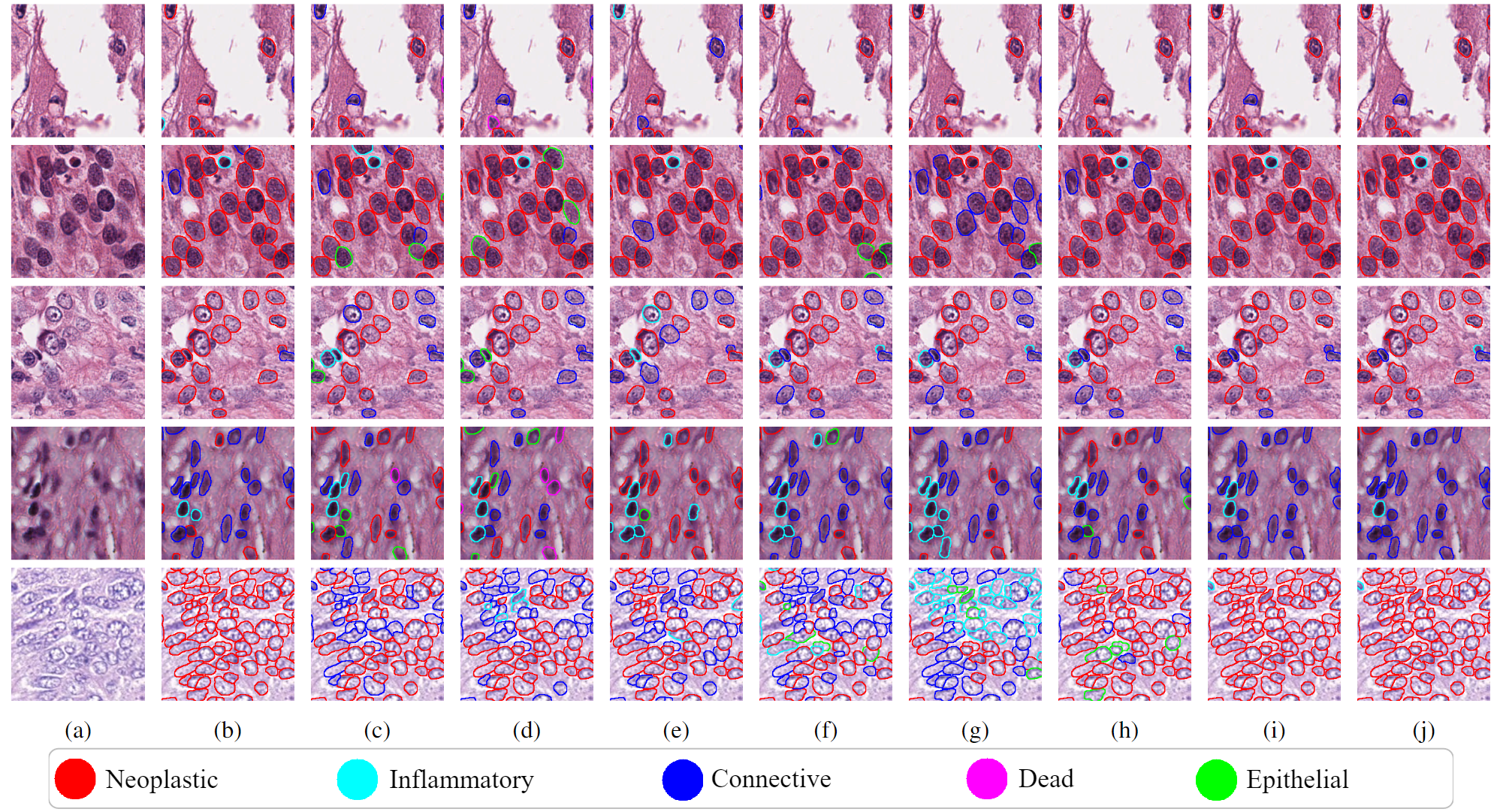
Results of different classification methods on histopathological patches of 40× in PanNuke. (a) Input patch. (b) SimCLR. (c) Moco. (d)Moco v2. (e) BYOL. (f) RCCNet. (g) ViT. (h) BiT. (i) Structured-Triplet. (j) Ground truth. We show that our method is more accurate than other methods and it outputs smoother classification results. It is intuitive in pathology images that nearby nuclei are more likely to belong to the same class.
|
| Data |
|
|
| Paper |
|
|
| Abstract |
|
The classification of nuclei in H&E-stained histopathological images is a fundamental step in the quantitative analysis of digital pathology. Most existing methods employ multi-class classification on the detected nucleus instances, while the annotation scale greatly limits their performance. Moreover, they often downplay the contextual information surrounding nucleus instances that is critical for classification. To explicitly provide contextual information to the classification model, we design a new structured input consisting of a content-rich image patch and a target instance mask. The image patch provides rich contextual information, while the target instance mask indicates the location of the instance to be classified and emphasizes its shape. Benefiting from our structured input format, we propose Structured Triplet for representation learning, a triplet learning framework on unlabelled nucleus instances with customized positive and negative sampling strategies. We pre-train a feature extraction model based on this framework with a large-scale unlabeled dataset, making it possible to train an effective classification model with limited annotated data. We also add two auxiliary branches, namely the attribute learning branch and the conventional self-supervised learning branch, to further improve its performance. As part of this work, we will release a new dataset of H&E-stained pathology images with nucleus instance masks, containing 20,187 patches of size 1024 x 1024, where each patch comes from a different whole-slide image. The model pre-trained on this dataset with our framework significantly reduces the burden of extensive labeling. We show a substantial improvement in nucleus classification accuracy compared with the state-of-the-art methods. |
| |
| Network Overview |
 |
|
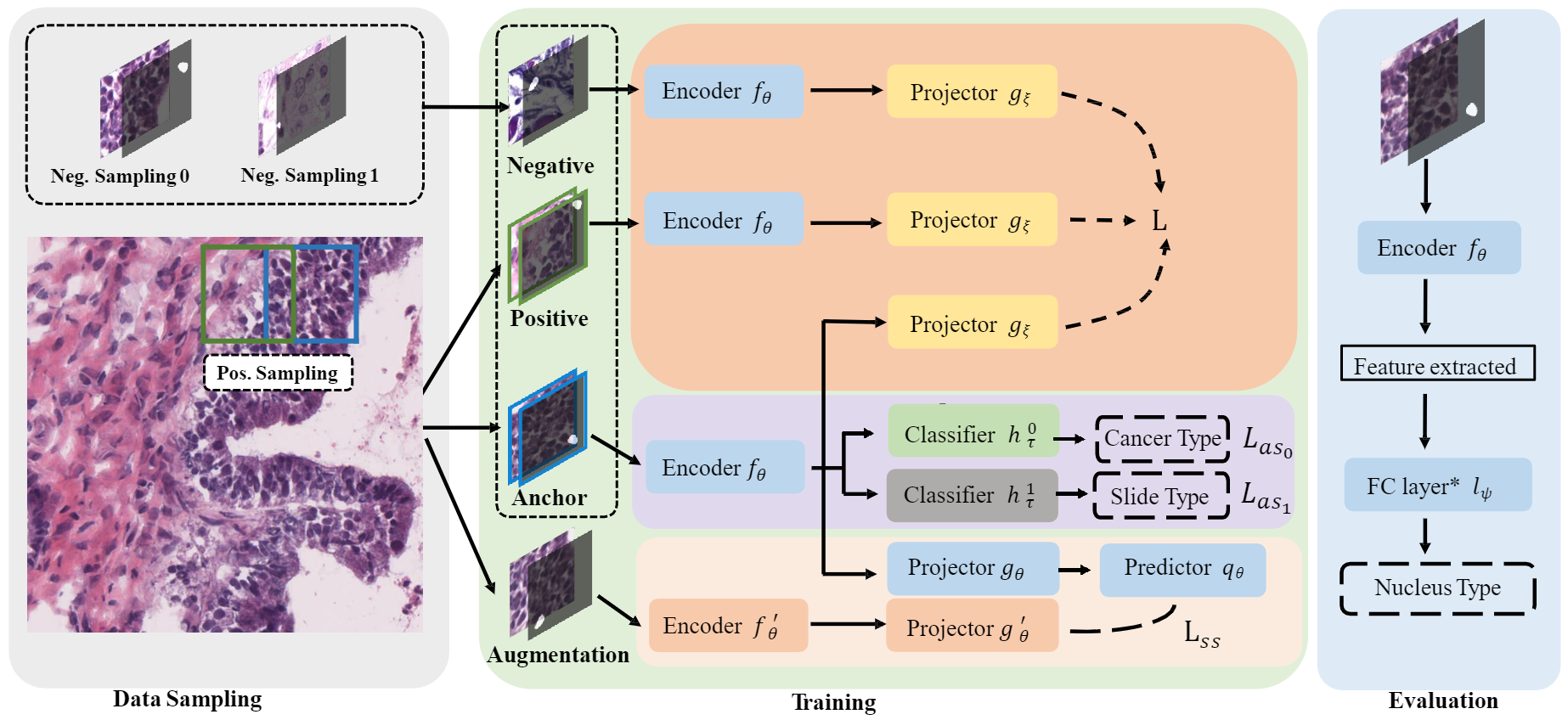
Pipeline of our framework. This framework trains a feature extraction model fθ with three branches on a large-scale unlabelled pathology dataset. The main body of the framework is Structured Triplet, a triplet learning branch with positive and negative sampling strategies tailored to our structured inputs. We also add two other auxiliary branches, the slide attribute learning branch and the conventional self-learning branch
|
| |
| ©Wenhua Zhang. Last update: Nov., 2022. |


